Method Article
Enhanced Gene Delivery and Expression using Intraosseous Injection of Chitosan Nanoparticles Encapsulated Adenine Base Editor Plasmids
* These authors contributed equally
In This Article
Summary
Here, we present a gene therapy approach that delivers ABE-coated chitosan directly to the bone marrow by intraosseous injection.
Abstract
The delivery of exogenous plasmids into experimental animals is crucial in biomedical research, including the investigation of gene functions, the elucidation of disease mechanisms, and the assessment of drug efficacy. However, the transfection efficiency of the current method is relatively low, and the introduction of plasmids for long-term gene expression may be affected by the immune system. To address these limitations, we developed and investigated a novel method that utilizes chitosan to encapsulate adenine base editor (ABE) plasmids and then directly deliver the complex to the bone marrow of mice by intraosseous injection. In this study, to target the CaMK II δ gene, which is closely related to osteoclast differentiation, we utilized chitosan to encapsulate ABE CaMK II δ plasmids. We directly injected the plasmid cargo into the bone marrow cavity by intraosseous injection. Results showed a high in vivo editing efficiency of 14.27% at the A1 locus and 10.69% at the A2 locus in recipient mice. This novel strategy is not only particularly suitable for diseases caused by abnormal osteoclast function but also holds significant potential for advancing the field of gene therapy.
Introduction
Gene therapy has emerged as a promising approach in the field of biomedical research1,2. It offers the potential to treat a variety of disorders by introducing foreign plasmids in experimental animals to modulate the expression of specific genes and study their therapeutic effects. However, conventional delivery methods have encountered some problems that limit their effectiveness and safety3. The concerns include low transfection efficiency, high biological damage, and low gene expression efficiency. Therefore, it is necessary to establish a novel delivery method for gene therapy that can overcome these limitations.
Chitosan is a natural polysaccharide with good biodegradability and biocompatibility4,5,6, which makes it easy to degrade in animal bodies without causing serious biological toxicity. It has been widely used in drug delivery due to its high drug embedding rate7,8, enhanced delivery efficiency, and reduced damage to animals.
ABE gene editing technology, which allows direct base pairs conversion of A-T to G-C, is promising in genetic and medical research. Compared to the current mainstream gene editing technology, ABE technology can achieve accurate single base mutation, thus reducing editing of non-target DNA sequences, reducing off-target effects9,10, and leading to zero DNA double-strand breaks11, which greatly reduces the risk of gene editing12. ABE technology is also highly biocompatible and is more suitable for disease treatment research.
Tail vein injection is a common method used for in vivo delivery of plasmids, especially in gene therapy. The target gene for this study is CaMK II δ, which is closely related to osteoclast differentiation13,14. The use of intraosseous injection instead of the tail vein allows the edited plasmid to enter the osteoclasts directly into the bone marrow. This direct transmission to the bone marrow increases the efficiency and stability of gene expression, which is advantageous for the treatment of diseases related to osteoclast dysfunction.
Here, we present a novel method that involves encapsulating the ABE plasmid with chitosan and then directly introducing the complex into mice by intraosseous injection. Through this method, we hope to pave the way for more effective gene therapy treatments for foreign plasmids entering organisms, especially diseases related to osteoclast dysfunction.
Protocol
All animal experiments described were approved by the Animal Health Committee of Anhui University on the Use and Care of Animals. In this study, ABE was generously donated by Professor Tian Chi (Shanghai University of Science and Technology, Shanghai, China ; Figure 1D).
1. ABE plasmid construction
- Design gRNA with the sequence TCCATGATGCACAGACAGGA where PAM is presented by GAC.
- According to the company's primer list guide, add the primer to the corresponding volume of ddH2O to achieve a concentration of 100 µM. Then, shake and mix thoroughly to ensure even distribution. For the annealing reaction, prepare a 10 µM solution by adding 10 µL of the stock primer solution to the new microcentrifuge tube, 90 µL of ddH2O, and mix well.
- Add 5 µL of upstream primer, 5 µL of downstream primer, and 40 µL of ddH2O in a microcentrifuge tube. To facilitate the annealing reaction, immerse the tube into a container filled with boiling water for 5 min. Ensure that the water level does not reach the opening of the tube to prevent water from entering and contaminating the primers. Cool down to room temperature to complete the annealing reaction.
- Add 2 µg pLKO5.sgRNA.EFS.tRFP (Figure 1D), 1 µL of ESP3I, 4 µL of buffer, and 33 µL of ddH2O into a microcentrifuge tube and allow to react in a constant temperature water bath at 37 °C for 45 min.
- Perform agarose gel electrophoresis. Prepare a 1% agarose gel by weighing 0.5 g agarose and pour it into a glass bottle. Add 50 mL of 1x TAE buffer into the glass bottle and shake to dissolve the agarose powder. Heat in the microwave oven for 2 min until completely melted. Add 5 µL of YeaRed Nucleic Acid Gel Stain, mix thoroughly, and pour into the gel mold; after the gel solidifies, start adding the samples.
NOTE: To prepare 1xTAE, use the reagents listed in Table 1. - Add 1x TAE to the electrophoresis tank and put in the configured agarose gel block, which should be immersed in TAE. Add 5 µL of 1x loading buffer to the digestion product from step 1.4 and mix. Add the sample to the gel. In addition, add 6 µL of marker to the gel as well.
- Run the electrophoresis program at a constant voltage of 110 V for 35 min. After electrophoresis, determine the position of the band in the sample according to the marker and cut the gel strip with a clean blade for gel recovery.
- Perform the gel recovery operation according to the gel maxi purification kit instructions. Set a water bath to 50 °C. Add the corresponding volume of PN solution to the tube containing the cut gel as per the kit. Poke the gel with a pipette tip to facilitate dissolution and then put it in the water bath. Shake the tube every once in a while and observe the dissolution process until the gel block is completely dissolved.
- Take a spin column, CA3, from the kit. Add 500 µL of BL solution from the kit into the spin column and centrifuge it at 2,000 x g for 1 min to equilibrate the column. After centrifugation, remove the waste liquid at the bottom of the collection tube and put the adsorption column back into the collection tube.
- Continue centrifugation at 2,000 x g for 1 min. Discard the waste liquid at the bottom of the centrifuge tube, add 600 µL of PN solution, and centrifuge 2,000 x g for 30 s; discard the waste liquid again. Repeat the operation and again discard the waste liquid.
- After centrifugation, replace the 2 mL collection tube with a new 1.5 mL tube. Open the cover and let dry in a ventilated place for 30 min to remove the ethanol completely. Then, add preheated 40 µL of ddH2O, centrifuge, and elute the required DNA.
NOTE: PN should be added with anhydrous ethanol according to the instructions. The use of preheated water can increase the yield. - Add 7 µL of annealing product from step 1.3, 3 µL of gel recovery product from step 1.11, 1 µL of T4 DNA Ligase, 2 µL of 10x T4 Ligase Buffer, and 7 µL of ddH2O into a microcentrifuge tube, and keep overnight at 16 °C to obtain the ligation product.
- Add 10 µL of ligation product and 70 µL of DH5α in a microcentrifuge tube on ice for 30 min. Set the water bath to 42 °C. After the ice bath, put the tube into the water bath for 90 s and then again on ice for 2 min.
- Add 600 µL of Luria-Bertani (LB) liquid medium to the tube and shake on a shaker for 30 min. At the same time, irradiate an LB solid medium plate inside a laminar flow cabinet for 30 min. After the incubation, evenly distribute 100 µL of the solution on the plate. After coating, place the plate upside down in a 37 °C bacterial incubator for culture.
NOTE: DH5α is usually stored at -80 °C. LB solid medium is prepared using the reagents listed in Table 2. LB liquid medium is prepared using the reagents listed in Table 3. - Select a single colony and expand the bacterial colony for 14 h on a fresh plate. Take a 50 mL centrifuge tube, add 50 mL of LB liquid medium, add 50 µL of ampicillin at a 1:1000 ratio, and mix well. Divide this into new tubes at 15 mL per tube.
- Check the culture plate for round white colonies with good growth. Select these with a 10 µL pipette and transfer them to the LB medium. Culture this in a shaker for 12 h and then send this sample for sequencing.
- Compare the sequencing results with the target gRNA to identify the strain with the matching sequence, Camk II δ-sgRNA, as the target strain, and then extract the plasmid.
- Perform plasmid extraction according to the instructions of the mini plasmid kit. Add 500 µL of BL to the adsorption column CP3, centrifuge for 1 min at 2,000 x g, and discard the waste liquid. Put the adsorption column back into the collection tube. Add 500 µL of bacterial solution and centrifuge at 4,000 x g for 10 min.
- Collect the flow through. Centrifuge again, and after removing the supernatant, add 250 µL of P1 to the centrifuge tube with bacterial precipitation and then transfer it to a microcentrifuge tube and vortex. Add 250 µL of P2 and gently invert for 6x-8x. Add 350 µL of P3 and again gently invert for 6x-8x. White flocculent precipitate appears.
- Centrifuge for 10 min at 2,000 x g. Transfer the supernatant to a new adsorption column CP3, centrifuge at 2,000 x g for 30 s, and remove the waste liquid. Add 600 µL of PW, centrifuge again at 2,000 x g for 30 s, and remove the waste liquid. Add 600 µL of PW again, centrifuge at 2,000 x g for 30 s, and remove the waste liquid.
- Place the adsorption column in the collection tube and centrifuge for 2 min at 2,000 x g to remove residual PW. Then, place the adsorption column in a clean microcentrifuge tube, allow it to dry for 30 min, and add 35 µL of preheated ddH2O to the middle of the adsorption film, then store the resulting product in -20 °C for subsequent tests.
NOTE: PW should be added with anhydrous ethanol according to the instructions. The use of preheated water can increase the yield.
2. Marrow cell extraction
- Select KM mice aged 8-9 weeks, male, and weighing about 30 g. Fix the mice in the left hand with the head facing down and inject 150 µL of 1% Pentobarbital sodium from the side of the abdomen with a syringe using the right hand. Confirm anesthesia by toe pinch.
- Perform cervical dislocation of mice, remove the tibia with surgical scissors and tweezers, and strip away the surrounding excess muscle tissue. Soak the removed tibia in PBS and wash, leaving only clean bones.
- Take 1 mL of PBS in a syringe to flush bone marrow cells from one end of the tibia until the bone is white.
- Collect cell suspension and centrifuge at 800 x g at 4 °C for 5 min. After removing the supernatant, lyse the red blood cells with 500 µL of red cell lysate. After 1 min, add 1 mL of separation buffer to terminate the red cell lysate. After centrifuging at 800 x g at 4 °C for 5 min, remove the supernatant and place the cells on ice for later use.
NOTE: If the red blood cells are completely lysed, the cells precipitate, and no red cells are visible. If the red cells can still be seen, they can be lysed again, but the incubation time should not be too long to avoid damage to other cells.
3. Chitosan transfection
- Chitosan preparation: Weigh 4 mg chitosan and dissolve in 20 mL of 0.2 mg/mL acetic acid solution. Adjust pH to 5.5 with 10 M NaOH. Take 500 µL of this solution and add it to individual microcentrifuge tubes.
- Add 2 µg, 3 µg, 4 µg, and 5 µg ABE plasmids to individual tubes and dissolve them in 500 µL of 30 mM Na2SO4. Mix 500 µL of chitosan solution and 500 µL of the plasmid.
- Incubate them in a water bath at 50-55 °C for 15 min. After mixing the two solutions well, vortex for 15-30 s and let stand for 30 min.
- Chitosan characterization: Analyze the diameters and zeta potentials of Chitosan using dynamic light scattering (DLS), with the concentration of chitosan maintained at 0.1 mg/mL in ddH2O. Run each sample 3x.
- Cast 0.1 mg/mL samples onto a silicon chip. Add 20 µL of the resuspended samples to 200-mesh grids and incubate at room temperature for 10 min. Stain the grids with 2% phosphotungstic acid for 3 min and remove the remaining liquid with filter paper. Observe with a transmission electron microscope.
- Nanoparticle encapsulation efficiency: Filter the supernatant with a 0.1 µm filter to remove the unprecipitated particles. Upload the plasmid and measure the concentration with a spectrophotometer at 260 nm. The calculated efficiencies of ABE Plasmid and gRNA plasmid were 84% ± 0.37% and 85% ± 0.53%, respectively.
- Inject 100 µL of chitosan embedded with plasmid into mice through the bone marrow cavity (step 5). After 7 days of injection, isolate the bone marrow cells and lyse the red blood cells as described in step 2.
- Add 2 mL of serum-containing DMEM medium to 6-well cell culture plates and add the bone marrow cells (approximately 1 x 106) suspended in 500 µL of serum-containing DMEM medium. Measure the fluorescence intensity under a fluorescence microscope. CaMK II δ-sgRNA plasmid has TagRFP red fluorescence when excited at 580 nm (Figure 2B).
NOTE: Serum-containing DMEM medium is prepared using the reagents listed in Table 4.
4. Flow cytometry
- Prepare different dosages of ABE plasmids: 2 µg, 3 µg, 4 µg, and 5 µg, and deliver to recipient mice as described in step 3. After 7 days, isolate the bone marrow cells from the mice as described in step 3.
- Collect the bone marrow cells and centrifuge at 800 x g at 4 °C for 5 min. Discard the supernatant and lyse the red blood cells with 500 µL of red blood cell lysate. After 1 min, terminate the reaction with 1 mL of separation buffer and count the number of cells with a cell counter.
- Transfer 1 x 106 cells into a new microcentrifuge tube and centrifuge with 800 x g at 4 °C for 5 min. After removing the supernatant, resuspend the cells with 500 µL of PBS and transfer them into a flow tube. Measure the efficiency by flow cytometry. Prepare a flow cytometry control group as described in step 2.
NOTE: Cell debris and dead cells with the lower FSC signals and located in the lower-left corner of the FSC versus SSC scatter plot (less than 400) were excluded (Figure 3). Because the plasmids have TagRFP red fluorescence, they can be sorted out.
5. Bone marrow cavity injection
- Select KM male mice aged 8 to 9 weeks old, weighing about 30 g. Anesthetize them with an intraperitoneal injection of 150 µL of 1% Pentobarbital sodium (step 2.1). Confirm the depth of anesthesia by toe pinch.
- Fix the anesthetized mouse in a supine position on the operating table and fix the front limb of the mouse with tape.
- Disinfect the posterior tibia of mice with an alcohol swab. Fill the plasmid to be injected in a 1mL syringe with a 26G needle. Remove air bubbles and prepare for injection.
- Touch the shin of the mouse and hold the shin of the mouse with your fingers. Determine the position of the injection needle so that the tibial shaft is penetrated from the tibial plateau at the mouse's knee joint. Rotate the injection needle parallel to the tibia, stab into the bone marrow cavity, and inject slowly (about 3 s) to minimize the damage to the mice.
- After the injection, slowly pull out the needle (about 3 s) and immediately use an alcohol swab to stop bleeding and disinfect the puncture site.
- Gently place the mice in a warm environment (20-26 °C), wait for the mice to recover from anesthesia, and observe their recovery.
6. Sanger sequencing
- After 7 days of injection, collect mouse bone marrow cells as described in step 2 and isolate genomic DNA using an extraction kit. Collect the cells by centrifuging at 2,000 x g for 1 min. Discard the supernatant, add 200 µL of GA solution, and vibrate with a vortex mixer. Add 20 µL of Proteinase K and 200 µL of GB, mix, and then place in a 70 °C water bath for 10 min.
- After the incubation, add 200 µL of anhydrous ethanol, shake for 15 s, and then put the mixed liquid into the supporting adsorption column. Centrifuge at 2,500 x g for 1 min, discard the waste liquid at the bottom of the tube and add 500 µL of GD solution. Centrifuge and discard the waste liquid, wash the pellet 2x with PW, and finally dry it, eluting it with 40 µL of pure water.
NOTE: PW should be added with anhydrous ethanol according to the instructions. - Amplify the target gene CaMKIIδ by PCR with the forward primer sequence gctaaggtgataaatgtggcact and the reverse primer sequence of ctagtgtgcgggccagattc. Prepare the PCR reaction by adding 25 µL of 2x buffer, 1 µL of dNTP Mix, 2 µL of forward primer, 2 µL of reverse primer, 1 µL of DNA Polymerase, 2 µL of template DNA, and 17 µL of ddH2O. Run the following PCR program: 95 °C for 3 min, 95 °C for 15 s, 53.7 °C for 30 s, 72 °C for 60 s, 12 °C for heat preservation.
- After PCR, prepare a 1% agarose gel and perform agarose gel electrophoresis at 110 V for 35 min (step 1.5, step 1.6). Cut the target gel area after electrophoresis and send for Sanger sequencing (Figure 4).
Results
In vitro plasmids were injected into mice by intraosseous injection (Figure 1A). The average particle size of the nanoparticles was about 202.9 nm, the potential was 2.77 mV, and the PDI was 0.22 (Figure 1B). Figure 1C shows the surface shape of the nanoparticles observed as spherical by electron microscopy. Figure 1D shows the plasmid map of the ABE vector and the plasmid map of the gRNA vector.
Plasmid transduction was observed under a fluorescence microscope. Figure 2A shows normal bone marrow cells. After 7 days of plasmid transfer into the mouse, the bone marrow cells of the mouse were observed under a fluorescence microscope. Compared with the control group, the transfection efficiency of the chitosan transfection group was significantly improved (Figure 2B). This indicates that chitosan as an embedding material can improve the efficiency of transduction plasmid in vitro.
After 7 days of delivery into the mice, the plasmid was fully expressed. Bone marrow cells from mice were extracted, and the transfection efficiency of bone marrow cells was detected by flow cytometry. The results showed that the transfection efficiency of cells directly transfected with ABE plasmid was 0.92% ± 0.02%. The transfection efficiency of bone marrow cells transfected with 2 µg ABE coated with chitosan was 40.80% ± 4.31%, that of bone marrow cells transfected with 3 µg ABE coated with chitosan was 47.20% ± 5.37%, that of bone marrow cells transfected with 4 µg ABE coated with chitosan was 51.20% ± 2.02%, and that of bone marrow cells transfected with 5 µg ABE coated with chitosan was 48.2% ± 7.39% (Figure 3B). The efficiency of the Chitosan transfection group was higher than that of the control group (Figure 3A). Flow cytometry results showed that the transduced plasmid could be stably and efficiently expressed in mice using the method described in this paper. From the perspective of comprehensive expression efficiency and material saving, the scheme of one-time transduction of 3 µg plasmid could be selected.
We extracted genomic DNA from bone marrow cells and amplified the target gene by PCR. After Sanger sequencing, the in vivo editing efficiency at A1 was 14.27% ± 0.35%, and that at A2 was 10.69% ± 0.30% (Figure 4).

Figure 1: Bone injection flow chart, nanoparticle characterization, and the plasmid vector map. (A) Procedure for intraosseous injection. (B) Particle size, Zeta potential, and PDI of nanoparticles formed by ABE plasmids embedded with chitosan. (C) Electron microscopic image of nanoparticles. (D) The left side shows the map of the ABE vector plasmid, and the right side shows the map of the gRNA vector plasmid. Please click here to view a larger version of this figure.
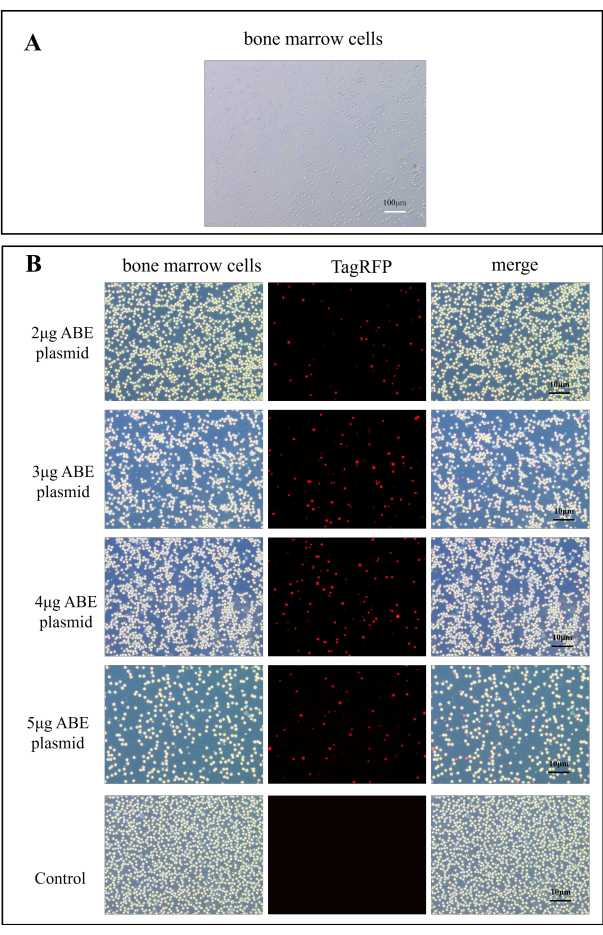
Figure 2: Fluorescence images for ABE plasmid transfection efficiency. (A) A100x image of bone marrow cells. (B) The fluorescence maps of bone marrow cells in the chitosan transfection group were obtained 7 days after transfection of 2 µg, 3 µg, 4 µg, and 5 µg ABE plasmid, compared to those in the control group after direct injection of ABE plasmid Please click here to view a larger version of this figure.
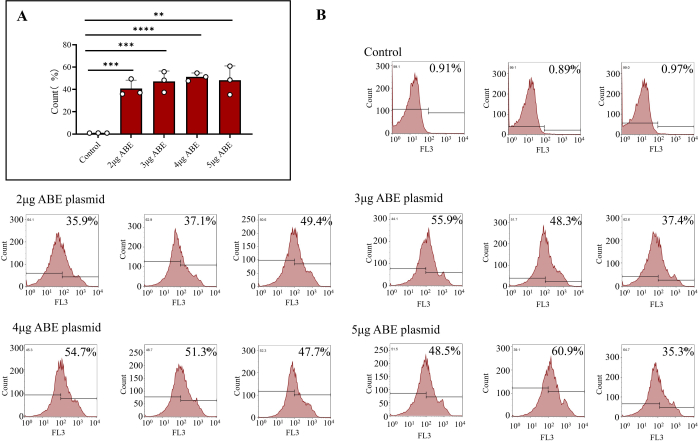
Figure 3: Flow cytometry results of transfected bone marrow cells. (A) Flow cytometry treated bone marrow cells transfected with 2 µg, 3 µg, 4 µg, and 5 µg ABE plasmids (Chitosan transfection group) and directly injected (control group). The results showed that the transfection efficiency of bone marrow cells directly injected with ABE plasmids was poor, while the transfection efficiency and transfection effect of the other four groups were higher. (B) Flow cytometry results include the transfection efficiency of the control group, and different ratios of ABE plasmid. Please click here to view a larger version of this figure.
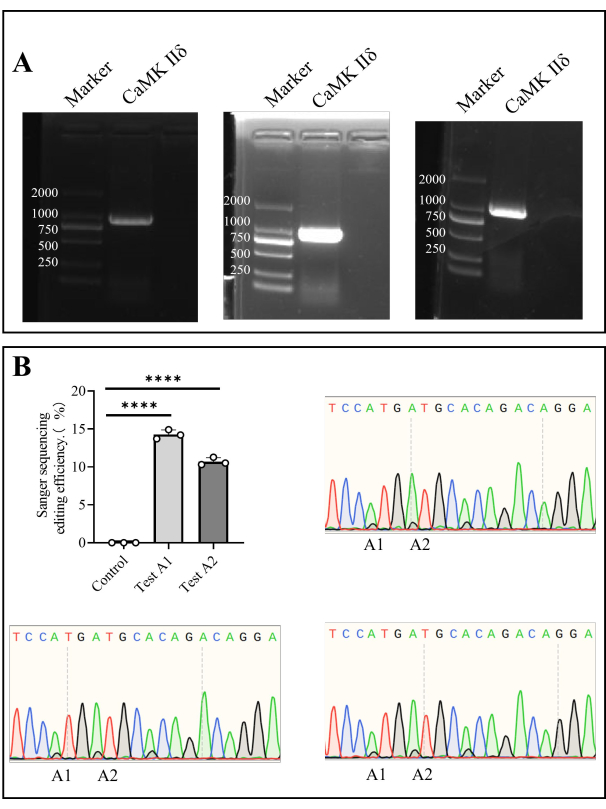
Figure 4: Sanger sequencing of Camk II δ editing efficiency in mice. (A) Agarose gel electrophoresis detection of Camk II δ. (B) The bar chart shows the Sanger sequencing result analysis of the editing efficiency of Camk II δ in mice, and the Sanger sequencing diagrams show the sequencing results. Please click here to view a larger version of this figure.
| Reagents | Concentration used |
| Ethylenediaminetetraacetic acid | 1.142 mL |
| Glacial acetic acid | 2 mL |
| Tris | 4.84 g |
Table 1: Reagents for preparing 1X TAE buffer
| Reagent | Volume |
| Agar | 7.5 g |
| Amp | Add 500 μL when sterilized and cooled to 70 °C |
| ddH2O | To 500 μL |
| Sodium chloride | 5 g |
| tryptone | 5 g |
| Yeast extract | 2.5 g |
Table 2: Reagents for preparing LB solid medium.
| Reagent | Volume |
| ddH2O | To 1 L |
| peptone | 10 g |
| Sodium chloride | 10 g |
| Yeast extract | 5 g |
Table 3: Reagents for preparing LB liquid medium.
| Reagent | Volume |
| DMEM | 450 mL |
| Fetal bovine serum | 5 mL |
| MEM | 5 mL |
| Penicillin/Streptomycin | 5 mL |
| β-mercaptoethanol | 500 μL |
Table 4: Reagents for preparing serum-containing DMEM medium.
Discussion
For biomedical research, the challenge in delivering exogenous plasmids to animals involves improving the efficiency of delivery and gene expression while simultaneously minimizing harm to animals to achieve the desired therapeutic effect15,16. We present a novel method of intraosseous injection of chitosan-mediated ABE plasmid delivery into the bone marrow cells of recipient mice. This strategy improves plasmid delivery efficiency and gene expression.
First, in this study, chitosan, offered as a nonviral vector system, was used as a method for plasmid delivery. Since it has been demonstrated that the transfection efficiency of the conventional method is relatively low, the introduction of plasmids for long-term gene expression may be affected by the immune system17,18. Chitosan, a natural polysaccharide, is renowned for its excellent biocompatibility, plasmid encapsulation ability, and low toxicity. Using chitosan as a delivery vehicle helps prevent potential immune-related effects during long-term expression of exogenous plasmid while also enhancing plasmid transfer efficiency19,20,21. For future research, we acknowledge the importance of evaluating the toxicity and potential bone marrow effects of chitosan nanoparticles when administered by intraosseous injection. We plan to incorporate these aspects into our future experimental design, including conducting comprehensive toxicity assessments and histopathological analyses. This will involve evaluating various parameters such as cell viability, inflammatory responses, and bone marrow morphology at different time points and dose levels. Additionally, we will consider using advanced imaging techniques and molecular biology methods to gain a deeper understanding of the safety profile and potential impacts on bone marrow health following treatment with chitosan nanoparticles.
Second, ABE is selected as a gene editor that can convert adenine nucleotides into guanine nucleotides without introducing double-stranded DNA breaks. Compared to CRISPR/Cas9, ABE offers higher precision by directly correcting a single base without inducing double-strand breaks, thereby reducing off-target effects and potential damage. However, when designing the target gRNA, careful consideration is required to prevent non-specific binding and reduce off-target effects22. We have implemented measures to reduce the risk, but complete avoidance remains challenging and needs more research.
Third, for in vivo delivery, we developed an intraosseous injection strategy. The intraosseous injection is a safe and effective method, as it has been reported in the system review by Betzler et al.23. The target gene in this study is Camk II δ, which plays a key role in the regulation of osteoclast differentiation13,14. Compared with the conventional tail vein injection method, the intraosseous injection can deliver the targeted Camk II δ plasmid directly and efficiently into the osteoclasts, thereby achieving the high in vivo editing efficiency of 14.27% at A1 locus, while 10.69% at A2 locus in recipient mice.
Moreover, it should be noted that the intraosseous injection is an operation requiring special attention. So, before the experiment, we used blue dye as practice to monitor if the injection is correctly injected into the bone marrow cavity of the mice. In addition, due to the possibility that recipient mice might suffer from the damage caused by intraosseous injection, it is recommended to choose mice weighing about 30 g. We emphasize the importance of several key steps in the procedure of intraosseous injection: avoid administering an excessive amount of anesthetic, ensure thorough disinfection both before and after injection to prevent infection, and rotate the needle to avoid blockages caused by the bone marrow. If blocked, refrain from forcing the fluid into the mouse's bone marrow cavity, which may potentially lead to the death of the mouse. The plasmid should be injected slowly, and it is crucial to monitor the mice's condition post-injection to ensure that no fatalities occur due to improper manipulation. Although we specified the mice weigh 30 g for successful intraosseous injection. In clinical settings, patient-specific variability must be considered, and our future studies, such as optimizing injection techniques for different weight ranges and exploring alternative delivery methods, are currently ongoing.
This method can be widely used in the editing of bone marrow-derived cells, including osteoblasts, erythroid cells, lymphocytes, hematopoietic cells, etc. The high precision and efficiency of the method allow researchers to use it not only for diseases caused by abnormal osteoclast function but also in the field of gene therapy for other blood disorders. For example, common blood diseases can also be well aligned with our treatment methods. Blood diseases caused by single base mutations24 can be treated with ABE gene editors, and intraosseous injection can also deliver plasmids directly to hematopoietic stem cells, thus achieving better therapeutic effects. However, the method exhibits limitations in its application for the editing and delivery processes of cell types of non-bone marrow origin, such as muscle and adipose cells.
Disclosures
The authors declare no competing interests.
Acknowledgements
This work was funded by the Natural Science Foundation of Anhui Province (2208085MC74, 2208085MC51) and the Scientific Research Foundation from the Education Department of Anhui Province, China (KJ2021A0055).
Materials
| Name | Company | Catalog Number | Comments |
| 0.2 ml PCR Tubes, Flat cap | LABSELECT | PT-02-C | |
| 1 mL syringe | Anhui Jiangnan medical equipment Co., LTD | / | |
| 1% Pentobarbital sodium | / | / | |
| 1.5 ml Microcentrifuge Tubes | LABSELECT | MCT-001-150 | |
| 10 × DNA Loading Buffer | Vazyme | P022-01 | |
| 10X T4 DNA Ligase Buffer 1 ml | TaKaRa Biotechnology(Dalian)Co.,LTD | 2011A | |
| 1250 μl Pipette Tip 102.1mm | LABSELECT | T-001-1250 | |
| 200 μl Pipette Tips 50.55mm | LABSELECT | T-001-200 | |
| 2x Phanta Max Buffera | Vazyme | P505-d1 | |
| 4 ? centrifuge | Thermo Fisher | 75002425 | |
| 50 ml Centrifuge tube | LABSELECT | T-012-50 | |
| 6-well Cell Culture Plates | LABSELECT | 11110 | |
| Agar | Sangon Biotech | A505255-0250 | |
| Amp | Abiowell | / | |
| Chitosan | Sangon Biotech | A600614-0500 | |
| Constant temperature culture shaker | Shanghai Zhicheng Analytical Instrument Manufacturing Co., LTD | ZWY-200D | |
| Countess Automated Cell Counter | Thermo Fisher Scientific | Countess II/II FL | |
| Countess Cell Counting Chamber Slides and Holder, disposable | Thermo Fisher Scientific | C10228 | |
| CutSmart Buffer | New England Biolabs | B7204SVIAL | |
| DH5α | General Biosystems | CS01010 | |
| DMEM | gibco | C11995500BT | |
| dNTP Mix | Vazyme | P505-d1 | |
| Esp3I enzyme | NEBiolabs | R0734S | |
| Ethylenediaminetetraacetic acid | VETEC (sigma-aldrich) | V900106 | |
| Fetal bovine serum | OriCell | FBSAD-01011-500 | |
| Flow cytometer | BD FACSCalibur | 342975 | |
| Flow tube | Beyotime Biotechnology | FFC005-1bag | |
| Fluorescence microscope | Leica | 427019 | |
| gel maxi purification kit | TIANGEN | DP210 | |
| Genomic DNA extraction kit | TIANGEN | DP304 | |
| Glacial acetic acid | China National Pharmaceutical Group Corporation | 10000218 | |
| GoldBand DL2,000 DNA Marker | YESEN | 10501ES60 | |
| Ice machine | shanghaizhengqiao | BNS-30 | laboratory reserved |
| ImunoSep Buffer | Precision Biomedicals Co.,LTD | 604050 | |
| Megafuge 8 Small Benchtop Centrifuge Series | Thermo Fisher Scientific | 75004250 | |
| MEM | Life Technology | 11140050 | |
| NanoDrop 2000 | Thermo Fisher Scientific, USA | ||
| NaOH | SIGAM | S5881-500G | |
| PBS | XiGene | XG3650 | |
| PCMV-SPRY-ABE8E vector | / | / | |
| pcr amplification apparatus | Thermo Fisher | AKC96300441 | |
| Penicillin/Streptomycin | Solarbio | P1400 | |
| peptone | Sangon Biotech | A505247-0500 | |
| Phanta Max Super-Fidelity DNA Polymerase | Vazyme | P505-d1 | |
| Red cell lysate | Beyotime | C3702 | |
| Sodium chloride | China National Pharmaceutical Group Corporation | 10019318 | |
| Sodium sulfate | aladdin | S433911 | |
| T-001-10 10μl Pipette Tips 31.65mm | LABSELECT | T-001-10 | |
| T4 DNA Ligase | TaKaRa Biotechnology(Dalian)Co.,LTD | 2011A | |
| Tris | BioFroxx | 1115KG001 | |
| tryptone | Sangon Biotech | A505250-0500 | |
| vortex mixer | sigma | Z258423 | |
| Water bath | shanghaiyiheng | DK-80 | |
| YeaRed Nucleic Acid Gel Stain | YESEN | 10203ES76 | |
| Yeast extract | BBI | A610961-0500 | |
| Zetasizer Nano | Malvern Panalytical | Zetasizer Nano ZS | |
| β-mercaptoethanol | Sigma | 444203 |
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved