Method Article
Live Cell Imaging with Time Lapse Photography to Study Epidermal Keratinocyte Proliferation Kinetics
In This Article
Summary
Here, we provide a method for live cell imaging analysis that can be used to manually track the lineages of passage 0 keratinocytes and that allows the collection of proliferation metrics, including cell division fate and cell cycle duration.
Abstract
Live-cell imaging is an evolving and somewhat challenging method to study keratinocyte behavior in vitro. Historically, keratinocyte division behavior was investigated via methods such as clonal analysis, immunostaining, and cell cycle analysis. None of these methods allow for the analysis of keratinocyte behavior at the single-cell level in real time. Over the past decade, groups have utilized live cell imaging to identify keratinocyte stem cells and committed progenitors without the need for labeling. Differences have been identified in each respective group's division behavior, rate of terminal differentiation, and cell cycle duration. Here, a method for keratinocyte live cell imaging with time-lapse photography and its analysis is described. Utilizing unpassaged keratinocytes is recommended for this method to most closely mimic in vivo behavior. Live cell imaging provides a unique ability to study stem cell and committed progenitor behavior at the single cell level and to determine division fates, cell cycle duration, as well as other proliferation metrics.
Introduction
The ability to visualize cell populations in vitro in real-time as they expand for extended periods of time is a unique benefit of live cell imaging. Live cell imaging allows for cell motility, migration, and proliferation to be assessed at the single-cell level. The goal of this protocol is to optimize the visualization of keratinocyte cultures via time-lapse photography, producing videos that can then be tracked manually to obtain granular data on cellular behavior.
Our focus is on proliferation kinetics. From the analysis of the live cell imaging videos, lineage trees can be elucidated, and the time between divisions (a proxy for cell cycle duration), as well as the proportions of divisions that lead to further division versus differentiation of the daughter cells can be assessed.
There is substantial donor-to-donor variability when dealing with primary keratinocytes and frequent failed attempts at cell propagation. Because of this, many investigators opt to use highly proliferative keratinocytes such as HaCaT cells or neonatal keratinocytes, often after they have undergone multiple passages in vitro1. Culturing primary keratinocytes from adult or aged skin for the purpose of lineage tracing can be challenging. However, there are issues with the use of passaged cells from cell lines or from male foreskin. Repeated passaging results in cells that are significantly different from their in vivo state2. Furthermore, HaCaT cells have been shown to react differently than primary keratinocytes in multiple assays3,4,5. To utilize cells that most closely resemble their in vivo counterparts, passage 0 keratinocytes from adult human donors are utilized. Keratinocyte stem cells and committed progenitors exhibit distinct differences in behavior, that allow colonies from either population to be distinguished via live cell imaging6. This relatively novel ability to visualize the behavior of single keratinocytes over the long term has been used in only a few previous studies using similar techniques6,7,8. This protocol outlines live cell imaging of primary keratinocytes utilizing the IncuCyte S3 Live-Cell Analysis System. From the lineage trees that are constructed, colony type can be determined (stem cell versus committed progenitor), as well as cell cycle duration and the proportion of differentiation divisions.
Protocol
This study was performed in accordance with the Declaration of Helsinki. All human tissue was obtained after approval by the University of California, San Francisco (UCSF) institutional review board (IRB), and consent was obtained for all tissue used.
1. Time lapse photography of passage 0 human keratinocytes
NOTE: This protocol is specific to the IncuCyte S3 and SX5.
- Ensure that appropriate approvals from the institution's human research committee have been obtained to use human tissue for the study.
- Isolate the keratinocytes from fresh skin as described previously9.
- Determine the seeding density for the assay. Run pilot studies with samples at multiple dilutions from different donors to understand the density of cells needed in order to ensure adequate colonies for study but prevent excessive colonies that result in colony overlap over the observation period. Run these pilots under identical experimental conditions (same plates, reagents, etc.), as these factors can alter results.
NOTE: A seeding density that is too low results in insufficient growth, whereas a seeding density that is too high results in the inability to accurately track cells due to the crowding of cells in the field.- For Passage 0 keratinocytes from neonatal foreskins under 48 h from a collection stored at 4 °C in collection media, ensure the seeding density is 1000-5000/cm2. For P1 keratinocytes, use 500-2000 cells/cm2. The further the passage, the lower the seeding density.
- Plate cells on the selected plate size.
NOTE: 96 well plates and microplates have a meniscus effect, resulting in non-uniform cell distribution, with much of the growth being outside of the field of the imager. Plates with larger wells (24 well plates for example) allow for more data to be captured. 24 well plates allow visualization of up to 36 fields, whereas 96 well plates allow visualization of up to only 5 fields. However, the number of fields captured on an instrument is finite, and more fields will use up more of the imager's capacity. Typically, a 24 well plate is used.- To achieve an even distribution of cells on the plate various techniques can be used. First, rather than individually seeding each well, aliquot the total media needed for all wells at a specific dilution into a microtube. Then, pipette the total number of cells needed for the aliquot to reach the desired density and gently invert the tube to homogenously distribute the cells.
- After plating, move the plate in a cross pattern three times (up-down, left-right) and then transfer to the incubator carefully.
CAUTION: If using a microplate, avoid the outer rows and columns of wells, as the time-lapse microscope generates heat when imaging and may cause evaporation of the media in those wells. The experimental wells should be grouped in the center of the plate and surrounded with wells containing PBS or other sterile fluid at maximum capacity to reduce evaporation/edge effects.
- Incubate cells for 24 h at 37 °C and 5% CO2 to allow for adherence.
- Change media after 24 h.
NOTE: Do not disturb the growing monolayer when suctioning. Always use warmed media (37 °C) and gently stream media into wells using the side wall. The media used here is Epilife/Supplement S7/Primocin. The use of antibiotics in media for long-term live cell imaging is recommended for this application. Extended culture in a machine being used for multiple concurrent experiments is at high risk for contamination. Penicillin and streptomycin are commonly used. - Open the incubator by pressing the large triangular button on the bottom left to open the tray when the light is green. Put the vessel in an open bay. Then, close the tray using the same button on the bottom left. Never open the tray when the button to open is red, as that means it is actively scanning, and the scan will be interrupted.
CAUTION: Make sure there are no scans beginning by looking at the screen on the controller module. It tells the time until the next scan and the length of the scan. If scanning has started, it will give the time until the scan is completed. - Open the application on the computer and log in using the appropriate user ID and password. Select Schedule.
- Press the + button in the top left corner underneath the schedule to add the plate to the schedule.
- Select Scan on Schedule, then click Next.
- Select New, then click Next.
NOTE: If this plate is identical to a previous or running experiment, Copy Previous or Copy Current can be used to expedite formatting the experiment. - Select Standard for scan type, then click Next.
NOTE: Image Lock is a proprietary 96 well plate that helps minimize the occurrence of loss of focus/image jumping, which can be helpful if these issues are faced. The other scan types are not particularly useful for lineage tracing. - For scan settings, select Adherent Cell-by-Cell using the phase channel at 10x objective, then click Next.
NOTE: As long as the phase channel is chosen, no analysis option needs to be selected at this point, as the analysis can be initiated after data is captured. - Select the vessel, then click Next. Most common vessels are compatible with the machine, although non-microplates may need special attachments to fit in the time-lapse microscope's bays.
- Select an empty bay to place the vessel, then click Next.
- Select the wells to be scanned as well as the images per well, then click Next. The estimated scan duration will be provided on the bottom left of the screen.
- Name the experiment using the desired naming convention. Create a plate map of the experiment for future reference, then hit Okay. Finally, click Next.
- Defer analysis until after data collection, as the cell-by-cell analysis must be initiated after scanning has been completed. Click Next.
- Finally, set the frequency of imaging. In order to reliably track mitoses, which happen over 30 min, scan at 20-min intervals for keratinocytes. Press Next and confirm the settings to begin the experiment.
NOTE: Scan times cannot overlap, and the manufacturers recommend that the machine is at rest for as long as it is scanning. With 20-min intervals that means 10 min maximum should be spent scanning. If 12 wells of a 24 well plate are being imaged at 36 views per well, it takes 7 min for the scan to complete, so always be mindful of the resources available for the experiment when planning.- There is a notification if there is inadequate time for the machine to rest, and if scan times overlap then the vessel cannot be added to the schedule. If the vessel cannot fit on the schedule on a regular interval, then click Reserve tray location and hit Next.
- Double click the schedule on the top of the screen and select the vessel in reserve then manually click to add images on open time slots. Alternatively, right-click once the schedule is open to delete all scan times scheduled and right-click to set new scan times for a scan group at a regular interval.
NOTE: This would interrupt all experiments currently on the machine until new times were set. Do not forget to hit the floppy disk icon to save or the red X to cancel changes to the schedule. Selecting the desired wells or views is a straightforward process, as the machine immediately provides the estimated scan time upon parameter selection. The scan duration depends on the type and brand of plate selected, as well as the number of wells or views to be scanned. This allows users to adjust the scan parameters to match the machine's capacity.
- For keratinocytes, change media every 48 h. To ensure correct plate orientation, wait for the first scan to complete each time a vessel is placed in the machine (time permitting). It is a simple mistake to make. Place the plate into the machine so that the letters denoting the rows are on the left side of the bay.
- To view scans, press the View button with the eye icon and then double-click the experiment. Consider changing media early if there is a sudden change in the color of media (phenol red-containing media becoming yellow due to acidification), if there are excess dead cells present (can interfere with the proliferation of live colonies), or if abnormal morphology is observed.
- Once all wells have become quiescent or reached confluence, export the experiment. Follow adult colonies for ~2 weeks and neonatal for ~10 days, at which time crowding causes diminishing returns for the analyses.
- To export, click the View tab and double-click the experiment under the list of recent scans (the default is for them to be ordered from the most recent backward). Click the Landscape icon with the arrow and wait for the export tool to launch.
- Click As Displayed, then click Next.
- Manually click each field of view to export, then click Next.
- Select whether a movie or series of images is desired, and then select the scan times to export. For keratinocyte lineage tracing, use the movie option and select all scans (there is a quick Select all icon with a dashed line to expedite this). Click Next.
- Export at 1 frame per second at maximum quality (set this using the bar next to quality). Click Next.
- Choose the target folder and file type. Name the file and then click Export.
NOTE: It can take a long time, depending on the speed of the internet connection and computer involved, to export videos. Be prepared to wait multiple days to export all video files for larger experiments. Having the capability to log into the time-lapse microscope remotely is extremely convenient.
2. Using time-lapse imaging to construct lineage trees and generate data sheets
- Open the video file using a media player. VLC media player is recommended.
- Scroll through the videos and identify growing colonies. With VLC, use the arrow key to skip frames forward and backward. Take a screenshot of the colony to be tracked and label the colony.
- Identify a colony of interest either at the end of or after some days of video recording, then rewind the video and identify the colony forming cell.
NOTE: This is where having the appropriate seeding density is important. If too many colonies develop next to each other, they expand and merge into one another making it impossible to accurately track cell divisions. - When a cell is about to divide, it appears to condense (Figure 1). Pause the video when the division occurs. Record the time of the division (a timestamp is in the bottom left corner). Screenshot this initial division and give it the same label as the colony. Record the divisions in a hand-drawn lineage diagram (see Representative Results). Continue tracking and documenting as many generations as possible.
NOTE: Automated cell tracking has/is being developed to expedite this process and more accurately track cells7. Even in low calcium culture conditions with minimal differentiation machine learning models currently lack the sensitivity to accurately track keratinocyte divisions. - Transcribe manual lineage tree into data sheets - so named "green sheets" because of the color in the spreadsheet (Supplementary File 1).
- Use the green sheets to calculate the proportions of proliferative and differentiation divisions, identify stem cell and committed progenitor colonies, and cell cycle duration (see representative results).
NOTE: The machine is capable of analyzing data via its own basic and cell-by-cell analyzers to run multiple other assays (confluence, scratch). However, this is beyond the scope of this protocol.
Results
Primary keratinocytes grow in a stereotyped fashion, which can be tracked via live cell imaging. VLC media player is used to survey recordings. The time until the first division is variable and can be multiple days depending upon the characteristics of the donor, such as age, health status, or the growth factors present in vitro environment. Upon initial seeding, keratinocytes have a small, rounded appearance (Figure 1). After seeding, the colony-forming keratinocytes typically become flattened (Figure 1). These flattened keratinocytes tend to be more mobile than their non-dividing counterparts (Supplementary Video 1). Immediately before dividing, the flattened keratinocyte appears to condense centrally (Figure 1).
With the exception of the initial division, 95% of keratinocytes that are going to divide (proliferative - P) do so within 48 h of the previous division6. Those that do not are considered terminally differentiated (D) (Figure 1)6. These differentiated cells stay adhered until the end of the observation period or until they lift off the plate and are removed at the subsequent media change. Cells that differentiate tend to expand over time, resulting in non-uniform morphology of the keratinocyte colony (Figure 1). Export the videos once the observation period concludes and begin analysis (Figure 2).
Colonies are documented using a standardized process. The video prefix the colony was located in (A1, A2…, B1, B2…) is utilized, followed by the colony number. For example, A2-6 would be video A2, colony 6. The analysis begins with lineage tracing. Fast-forward to the end of a video to identify colonies, then rewind to the beginning of the observation period to identify the original colony-forming cell. Track the time stamps of all divisions as they occur and create a branched diagram by hand, tracking as many generations as can accurately be done (Figure 3). Eventually it is no longer possible to accurately track cell divisions due to the cellular density (this usually happens around generation 5-7, depending on whether the colony originated from a stem cell versus a committed progenitor). In later generations, the colony often merges with another colony, or the colony goes off-screen. At this point, mark the last trackable cell as a U (untraceable). Always take a screenshot of the colony being tracked (this can be done using the snapshot feature on VLC) and mark the colony using the nomenclature described above. Make sure to label the screenshot with standardized nomenclature, including the specific video analyzed and the colony number that can be matched to the screenshot.
Once the branch diagram has been constructed, data can be transferred to a spread sheet or "green sheet" (Supplementary File 1). The green sheet contains the same label for each colony tracked via branch diagram. To easily identify the generations, color highlights are alternated between generations. Time 1 of generation 1 refers to the period from plating the cells to placing them on the time-lapse microscope. In Supplementary File 1, time 1 of generation 1 is 24 h, as the cells are placed on the machine 24 h after plating. Time 2 represents the duration until the cell undergoes its first division. Recall that the time stamp in the videos does not account for the additional 24 h so the hours need to be manually added in each generation. When transcribing data from the branch diagram to the green sheet, always add 24 h to the time of divisions as the time stamp provided by the machine does not take into account the time before the recording starts. ΔT is the cell cycle duration. Most studies6,8 do not include the ΔT of the first generation in analyses, as it is always much longer than that of subsequent generations and extremely variable, resulting in skewed analyses.
Once the green sheet is constructed, one can determine which colonies originate from stem cells and which originate from committed progenitors. Colonies that show predominantly proliferative divisions (Figure 1) and last until the end of the observation period are considered stem cell colonies6. Colonies that terminally differentiate (Figure 1) during the observation period are considered committed progenitor colonies6. The mean ΔT of stem cell colonies and committed progenitor colonies, respectively, can then be calculated. By dividing the proportion of D divisions over total divisions, one can calculate the proportion of differentiation divisions, either with all pooled divisions of stem cell / committed progenitor colonies, respectively or by generation. The green sheet is a useful way to organize data to obtain information efficiently and it evolves depending on what data and statistics are needed for the goals of the study.
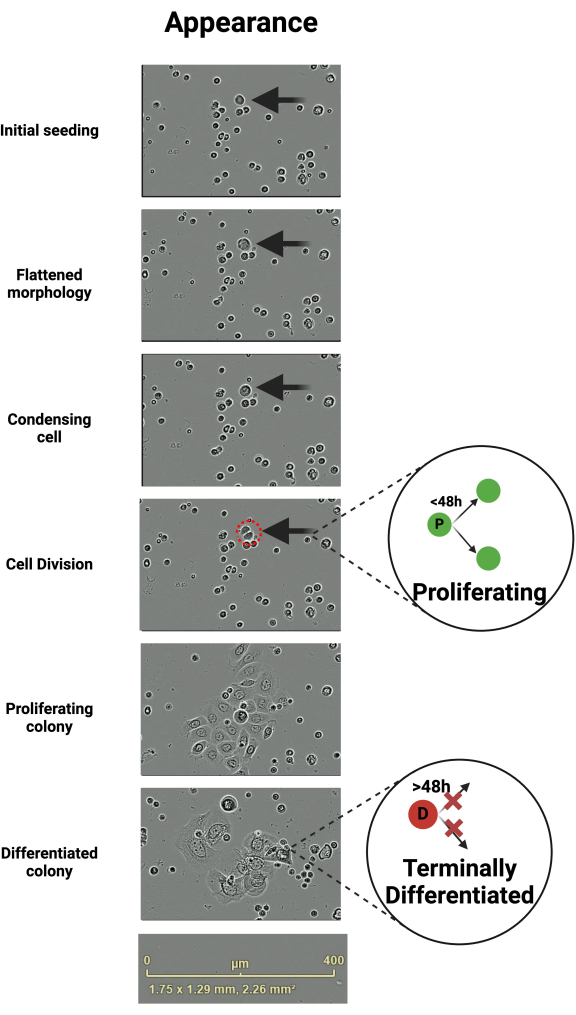
Figure 1: Changes in keratinocyte morphology leading up to cell division and division terminology. Keratinocytes progress through a stereotypic sequence of morphological changes leading up to cell division. This figure depicts a keratinocyte undergoing this sequence of events over time. Individual cells that are about to divide are motile and initially take on a flattened morphology and become condensed centrally immediately prior to dividing. Daughter cells that will continue to proliferate divide within 48 h of being generated otherwise they are considered terminally differentiated. Scale bar 400 µm. Created in BioRender. Ghadially, R. (2025) https://BioRender.com/k40e714 Please click here to view a larger version of this figure.
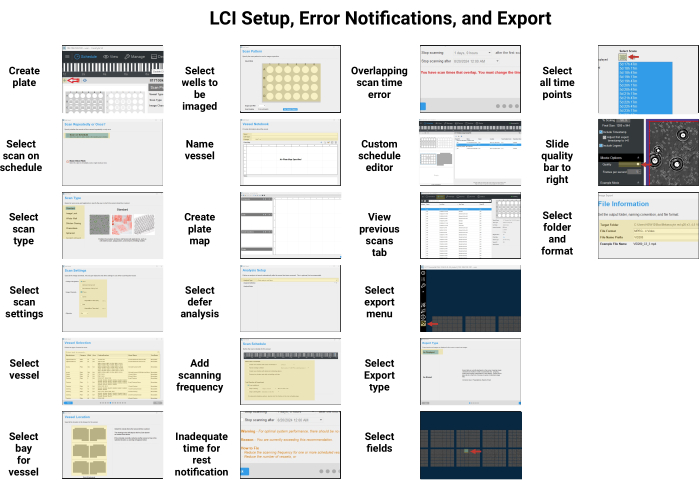
Figure 2: Exporting data from the time-lapse microscope. A guide on how to export data from the machine. Created in BioRender. Ghadially, R. (2024) BioRender.com/x99d452 Please click here to view a larger version of this figure.
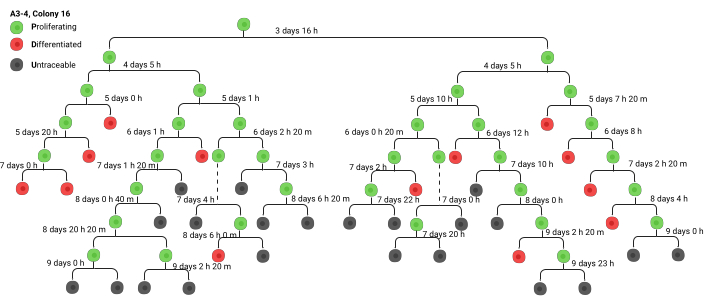
Figure 3. Precursor branch diagram (lineage tree) to inform the creation of green sheets. An example of a lineage tree, typically hand drawn. Abbreviations: h: hours, m: min. Created in BioRender. Ghadially, R. (2025) https://BioRender.com/wbt7z8x Please click here to view a larger version of this figure.
Supplementary File 1. Example data sheets (green sheets) of unpublished live cell imaging data. An example green sheet containing unpublished data. Please click here to download this File.
Supplementary Video 1. Example keratinocyte live cell imaging video. A live cell imaging video with suitable cell density to track keratinocyte colonies Please click here to download this File.
Discussion
Live cell imaging of keratinocytes is a label-free method to track the division behavior of stem cells and committed progenitors. Given that maintenance of the epidermis is dependent on the proliferation kinetics of stem cells and committed progenitors10, having a granular understanding of the changes in these keratinocyte populations and how they are affected in various conditions facilitates the development of therapies to ameliorate defects that are discovered.
Ultimately, lineage tracing via live cell imaging hinges upon obtaining usable data. The colony forming units need to be clearly visible in the videos generated by time-lapse photography. With freshly isolated passage 0 cells, obtaining a true clonal density is difficult. Only 3%-4% of plated keratinocytes ultimately form colonies11. Too many cells may make it impossible to track divisions or even identify the initial colony-forming cell. Too few cells, and colonies may not form. Similarly, anything that obscures the visual field makes it impossible to track cells via lineage tracing. Using fibroblast feeders may make it difficult to track divisions as fibroblasts obscure the visual field. Similarly, precipitates, cellular debris, and even condensation on the lid of the plate can obscure the growing colonies. Even something as small as not ensuring that the microplate is securely inserted into the selected bay may ultimately result in a failed experiment, as the image will not be focused on the cells. This is why it is very important to monitor the colonies daily while they are growing in the machine and always stay until after the first scan, whenever the microplate is removed from the machine for media changes, to ensure the cells are being captured by the time-lapse photography. Another issue that debris may cause is image jumping. Typically, the microscope has fixed visual fields that do not move, but when debris accumulates, or cell density gets too high, the machine may lose its field of view and jump to a different part of the plate. To avoid this, the manufacturers of the machine developed a special type of 96-well plate with a grid that prevents loss of focus.
The machine generates heat while capturing images. It is important to reduce the temperature of the incubator containing the microscope to 36.5 °C for the device to equilibrate to 37 °C when capturing images. When using plates with smaller surface areas (96 well plates), consider the additional heat generated. Do not use the peripheral wells for experiments (first and last rows/columns) and, consider maximally filling the wells surrounding experimental wells with other fluid (sterile PBS), and use breathable tape to reduce evaporative loss of media. There are plates marketed that could be investigated to minimize edge effects12. However, the aforementioned methods have allowed the use of recommended compatible microplates.
When analyzing videos, investigators need to be cognizant of abnormal division patterns from their colonies. For instance, if there are no divisions within a view for 10 days and then there is rampant colony formation originating from the edge of the field of view, it is unlikely to be the first division of a new colony. It is likely that a growing colony from a surrounding visual field encroached on the portion of the plate captured rather than being a novel colony. This can be verified by logging into the software suite, which can depict the entire plate (containing all recorded views) at specific time points and show the migration of cells from one field of view to another.
The primary limitation of this method is its labor-intensive nature. Also, tracking cell divisions past the 5th generation is difficult and requires hours of work, rewatching videos to accurately capture what is dividing and when. There are multiple deep learning automated cell tracking algorithms being developed, which will eventually result in purely AI-based analysis in the coming years7,13,14. Until then, manual tracking, such as detailed in this protocol, is a feasible method to develop lineage data.
Disclosures
None.
Acknowledgements
This work was supported by Merit Review Award Number I01 CX001816 from the United States (U.S.) Department of Veterans Affairs Clinical Sciences R&D (CSRD) Service. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. We thank Dr. Michael Rosenblum for providing us access to his time-lapse microscope to conduct our experiments.
Materials
| Name | Company | Catalog Number | Comments |
| 96 Well Imagelock plate | Sartorius | BA-04856 | Suggested microplate compatible with machine if using a 96 well plate. |
| 24 well plate | Corning | 3524 | Suggested microplate compatible with machine if using a 24 well plate. |
| Amphotericin B, 50 mL | Corning | 30-003-CF | Dilute to 5x (comes in 100x stock) for 5x PSA - 1x for media changes |
| Epilife, 50 mL | Gibco | MEP1500CA | Add S7, consider primocin |
| IncuCyte S3 | Sartorius | 4637 | Imager (Zoom/SX5 acceptable alternatives) |
| Penicillin/Streptomycin, 100 mL | Corning | 30-002-Cl | Dilute to 5x (comes in 100x stock) |
| Primocin | Invivogen | ant-pm-05 | 1 mL per 500 mL media |
| Supplement S7 | Gibco | S0175 | Added to epilife |
References
- Mateu, R., et al. Functional differences between neonatal and adult fibroblasts and keratinocytes: Donor age affects epithelial-mesenchymal crosstalk in vitro. Int J Mol Med. 38 (4), 1063-1074 (2016).
- Handl, J., Čapek, J., Majtnerová, P., Báčová, J., Roušar, T. The effect of repeated passaging on the susceptibility of human proximal tubular HK-2 cells to toxic compounds. Physiol Res. 69 (4), 731-738 (2020).
- Moran, M. C., et al. Characterization of human keratinocyte cell lines for barrier studies. JID Innov. 1 (2), 100018(2021).
- Seo, M. -D., Kang, T. J., Lee, C. H., Lee, A. -Y., Noh, M. HaCaT keratinocytes and primary epidermal keratinocytes have different transcriptional profiles of cornified envelope-associated genes to T helper cell cytokines. Biomol Ther. 20 (2), 171-176 (2012).
- Jahn, M., et al. Different immortalized keratinocyte cell lines display distinct capabilities to differentiate and reconstitute an epidermis in vitro. Exp Dermatol. 33 (1), e14985(2024).
- Xiao, T., et al. Short cell cycle duration is a phenotype of human epidermal stem cells. Stem Cell Res Ther. 15 (1), 76(2024).
- Hirose, T., Kotoku, J., Toki, F., Nishimura, E. K., Nanba, D. Label-free quality control and identification of human keratinocyte stem cells by deep learning-based automated cell tracking. Stem Cells. 39 (8), 1091-1100 (2021).
- Roshan, A., Murai, K., Fowler, J., Simons, B. D., Nikolaidou-Neokosmidou, V., Jones, P. H. Human keratinocytes have two interconvertible modes of proliferation. Nat Cell Biol. 18 (2), 145-156 (2016).
- Abegaze, B., Ijeh, N., Vittimberga, B., Ghadially, R. Generating primary cultures for the purpose of keratinocyte live cell imaging. J Vis Exp. , In Press (2024).
- Piedrafita, G., et al. A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nat Commun. 11 (1), 1429(2020).
- Ścieżyńska, A., et al. Isolation and culture of human primary keratinocytes-a methods review. Exp Dermatol. 28 (2), 107-112 (2019).
- Mansoury, M., Hamed, M., Karmustaji, R., Al Hannan, F., Safrany, S. T. The edge effect: A global problem. The trouble with culturing cells in 96-well plates. Biochem Biophys Rep. 26, 100987(2021).
- Malin-Mayor, C., et al. Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations. Nat Biotechnol. 41 (1), 44-49 (2023).
- Waliman, M., et al. Automated cell lineage reconstruction using label-free 4D microscopy. Genetics. 228 (2), iyae135(2024).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved